Prokaryote

A prokaryote (/proʊˈkærioʊt, -ət/; less commonly spelled procaryote)[1] is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles.[2] The word prokaryote comes from the Ancient Greek πρό (pró), meaning 'before', and κάρυον (káruon), meaning 'nut' or 'kernel'.[3] In the earlier two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. However in the three-domain system, based upon molecular phylogenetics, prokaryotes are divided into two domains: Bacteria and Archaea. A third domain, Eukaryota, consists of organisms with nuclei.
Prokaryotes evolved before eukaryotes, and lack nuclei, mitochondria, and most of the other distinct organelles that characterize the eukaryotic cell. Some unicellular prokaryotes, such as cyanobacteria, form colonies held together by biofilms, and large colonies can create multilayered microbial mats. Prokaryotes are asexual, reproducing via binary fission, although horizontal gene transfer may take place.
Molecular phylogenetics has provided insight into the evolution and interrelationships of the three domains of life. The division between prokaryotes and eukaryotes reflects two very different levels of cellular organization; only eukaryotic cells have an enclosed nucleus that contains its DNA, and other membrane-bound organelles including mitochondria. More recently, the primary division has been seen as that between Archaea and Bacteria, since the eukaryotes are part of the archaean clade and have multiple homologies with other Archaea.
Structure
[edit]The cellular components of prokaryotes are not enclosed in membranes within the cytoplasm, like eukaryotic organelles. Bacteria have microcompartments, quasi-organelles enclosed in protein shells such as encapsulin protein cages,[4][5] while both bacteria and some archaea have gas vesicles.[6]
Prokaryotes have simple cell skeletons. These are highly diverse, and contain homologues of the eukaryote proteins actin and tubulin. The cytoskeleton provides the capability for movement within the cell.[7]
Most prokaryotes are between 1 and 10 μm, but they vary in size from 0.2 μm in Thermodiscus spp. and Mycoplasma genitalium to 750 μm in Thiomargarita namibiensis.[8][9]
Bacterial cells have various shapes, including spherical or ovoid cocci, e.g. Streptococcus, Staphylococcus; cylindrical bacilli; spiral bacteria (twisted rods); and the comma-shaped vibrio.[10] Archaea are mainly simple ovoids, but Haloquadratum is flat and square.[11]
| Element | Description |
|---|---|
| Flagellum (not always present) | Long, whip-like protrusion that moves the cell. |
| Cell membrane | Surrounds the cell's cytoplasm, regulates flow of substances in and out. |
| Cell wall (except in Mollicutes, Thermoplasma) | Outer covering that protects the cell and gives it shape. |
| Cytoplasm | A watery gel that contains enzymes, salts, and organic molecules. |
| Ribosome | Structure that produces proteins as specified by DNA. |
| Nucleoid | Region that contains the prokaryote's single DNA molecule. |
| Capsule (only in some groups) | Glycoprotein covering outside the cell membrane. |
Reproduction and DNA transfer
[edit]Bacteria and archaea reproduce through asexual reproduction, usually by binary fission. Genetic exchange and recombination occur by horizontal gene transfer, not involving replication.[13] DNA transfer between prokaryotic cells occurs in bacteria[14] and archaea.[15]
Gene transfer in bacteria
[edit]In bacteria, gene transfer occurs by three processes. These are virus-mediated transduction;[14] conjugation;[16] and natural transformation.[17]
Transduction of bacterial genes by bacteriophage viruses appears to reflect occasional errors during intracellular assembly of virus particles, rather than an adaptation of the host bacteria. There are at least three ways that it can occur, all involving the incorporation of some bacterial DNA in the virus, and from there to another bacterium.[14]

Conjugation involves plasmids, allowing plasmid DNA to be transferred from one bacterium to another. Infrequently, a plasmid may integrate into the host bacterial chromosome, and subsequently transfer part of the host bacterial DNA to another bacterium.[18]
Natural bacterial transformation involves the transfer of DNA from one bacterium to another through the water around them. This is a bacterial adaptation for DNA transfer, because it depends on the interaction of numerous bacterial gene products.[17] The bacterium must first enter the physiological state called competence; in Bacillus subtilis, the process involves 40 genes.[19] The amount of DNA transferred during transformation can be as much as a third of the whole chromosome.[20][21] Transformation is common, occurring in at least 67 species of bacteria.[22]
Gene transfer in archaea
[edit]Among archaea, Haloferax volcanii forms cytoplasmic bridges between cells that transfer DNA between cells,[15] while Sulfolobus solfataricus transfers DNA between cells by direct contact. Exposure of S. solfataricus to DNA damaging agents induces cellular aggregation, perhaps enhancing homologous recombination to increase repair of damaged DNA.[23]
Colonies and biofilms
[edit]
Prokaryotes are strictly unicellular, but most can form stable aggregate communities in biofilms.[25] Bacterial biofilms are formed by the secretion of extracellular polymeric substance (EPS).[26] Myxobacteria have multicellular stages in their life cycles.[27] Biofilms may be heterogeneous and structurally complex and may attach to solid surfaces, or exist at liquid-air interfaces. Bacterial biofilms are often made up of microcolonies (dome-shaped masses of bacteria and matrix) separated by channels through which water may flow easily. Microcolonies may join together above the substratum to form a continuous layer. This structural complexity, combined with the easing of oxygen limitation by movement of medium throughout the biofilm, functions as a simple circulatory system,[28] approaching multicellular organisation.[29] Differential cell expression, collective behavior, signaling (quorum sensing), programmed cell death, and discrete biological dispersal events all seem to point in this direction.[30][31] Bacterial biofilms may be 100 times more resistant to antibiotics than free-living unicells, making them difficult to remove from surfaces they have colonized.[32]
Environment
[edit]
Prokaryotes have diversified greatly throughout their long existence. The metabolism of prokaryotes is far more varied than that of eukaryotes, leading to many highly distinct prokaryotic types. For example, in addition to using photosynthesis or organic compounds for energy, as eukaryotes do, prokaryotes may obtain energy from inorganic compounds such as hydrogen sulfide. This enables prokaryotes to thrive in harsh environments as cold as the snow surface of Antarctica, studied in cryobiology, or as hot as undersea hydrothermal vents and land-based hot springs.
Prokaryotes live in nearly all environments on Earth. Some archaea and bacteria are extremophiles, thriving in harsh conditions, such as high temperatures (thermophiles) or high salinity (halophiles).[33] Some archaeans are methanogens, living in anoxic environments and releasing methane.[2] Many archaea grow as plankton in the oceans. Symbiotic prokaryotes live in or on the bodies of other organisms, including humans. Prokaryotes have high populations in the soil, in the sea, and in undersea sediments. Soil prokaryotes are still heavily undercharacterized despite their easy proximity to humans and their tremendous economic importance to agriculture.[34]
Evolution
[edit]The first organisms
[edit]

A widespread current model of the origin of life is that the first organisms were prokaryotes. These may have evolved out of protocells, while the eukaryotes evolved later in the history of life.[36] An alternative model is that extant prokaryotes evolved from more complex eukaryotic ancestors through a process of simplification.[37][38][39]
Another view is that the three domains of life arose simultaneously, from a set of varied cells that formed a single gene pool.[40][41]
The oldest known fossilized prokaryotes were laid down approximately 3.5 billion years ago, only about 1 billion years after the formation of the Earth's crust. Eukaryotes only appear in the fossil record later, and may have formed from endosymbiosis of multiple prokaryote ancestors. The oldest known fossil eukaryotes are about 1.7 billion years old. However, some genetic evidence suggests eukaryotes appeared as early as 3 billion years ago.[42]
Phylogeny
[edit]According to the 2016 phylogenetic analysis of Laura Hug and colleagues, using genomic data on over 1000 organisms, the relationships among prokaryotes are as shown in the tree diagram.[43]

Classification
[edit]Taxonomic history
[edit]The distinction between prokaryotes and eukaryotes was established by the microbiologists Roger Stanier and C. B. van Niel in their 1962 paper The concept of a bacterium (though spelled procaryote and eucaryote there).[44] That paper cites Édouard Chatton's 1937 book Titres et Travaux Scientifiques[45] for using those terms and recognizing the distinction.[46] One reason for this classification was so that the group then often called blue-green algae (now cyanobacteria) would not be classified as plants but grouped with bacteria.[44]
In 1977, Carl Woese proposed dividing prokaryotes into the Bacteria and Archaea (originally Eubacteria and Archaebacteria) because of the major differences in the structure and genetics between the two groups of organisms. Archaea were originally thought to be extremophiles, living only in inhospitable conditions such as extremes of temperature, pH, and radiation but have since been found in all types of habitats. The resulting arrangement of Eukaryota (also called "Eucarya"), Bacteria, and Archaea is called the three-domain system, replacing the traditional two-empire system.[47][48]
As distinct from eukaryotes
[edit]The division between prokaryotes and eukaryotes has been considered the most important distinction or difference among organisms. The distinction is that eukaryotic cells have a "true" nucleus containing their DNA, whereas prokaryotic cells do not have a nucleus.[49]
Both eukaryotes and prokaryotes contain large RNA/protein structures called ribosomes, which produce protein, but the ribosomes of prokaryotes are smaller than those of eukaryotes. Mitochondria and chloroplasts, two organelles found in many eukaryotic cells, contain ribosomes similar in size and makeup to those found in prokaryotes.[50]
The genome in a prokaryote is held within a DNA/protein complex in the cytosol called the nucleoid, which lacks a nuclear envelope. The complex contains a single circular chromosome, a cyclic, double-stranded molecule of stable chromosomal DNA, in contrast to the multiple linear, compact, highly organized chromosomes found in eukaryotic cells.[51] In addition, many important genes of prokaryotes are stored in separate circular DNA structures called plasmids.[52] Like eukaryotes, prokaryotes may partially duplicate genetic material, and can have a haploid chromosomal composition that is partially replicated.[53]
| Domain | Nucleus | Organelles | Reproduction |
|---|---|---|---|
| Prokaryotes | None, DNA is free in cytoplasm | Few | Asexual, with horizontal gene transfer |
| Eukaryotes | DNA in nucleus | Membrane-bound organelles, inc. endoplasmic reticulum, mitochondria, chloroplasts | Sexual reproduction with haploid gametes |
Prokaryotes lack mitochondria and chloroplasts. Instead, processes such as oxidative phosphorylation and photosynthesis take place across the prokaryotic cell membrane.[54] However, prokaryotes do possess some internal structures, such as prokaryotic cytoskeletons.[55][56] It has been suggested that the bacterial phylum Planctomycetota has a membrane around the nucleoid and contains other membrane-bound cellular structures.[57] However, further investigation revealed that Planctomycetota cells are not compartmentalized or nucleated and, like other bacterial membrane systems, are interconnected.[58]
Prokaryotic cells are usually much smaller than eukaryotic cells. Therefore, prokaryotes have a larger surface-area-to-volume ratio, giving them a higher metabolic rate, a higher growth rate, and as a consequence, a shorter generation time than eukaryotes.[59]
Eukaryotes as Archaea
[edit]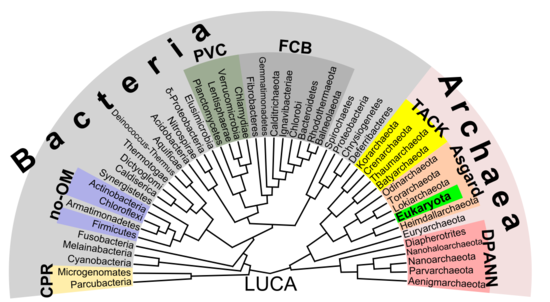
There is increasing evidence that the roots of the eukaryotes are to be found in (or at least next to) the archaean Asgard group, perhaps Heimdallarchaeota (an idea which is a modern version of the 1984 eocyte hypothesis, eocytes being an old synonym for Thermoproteota, a taxon to be found nearby the then-unknown Asgard group).[60] For example, histones which usually package DNA in eukaryotic nuclei, have been found in several archaean groups, giving evidence for homology.[61] The non-bacterial group comprising archaea and eukaryota was called Neomura by Thomas Cavalier-Smith in 2002, on the view that these form a clade.[62]
| Domain | Histones | ATP synthase | DNA replication |
|---|---|---|---|
| Archaea, inc. Eukaryota | Similar in these two groups, implying homology | ||
| Bacteria | (missing) | Present in a very different form | |
Unlike the above assumption of a fundamental split between prokaryotes and eukaryotes, the most important difference between biota may be the division between Bacteria and the rest (Archaea and Eukaryota).[60] DNA replication differs fundamentally between the Bacteria and Archaea (including that in eukaryotic nuclei), and it may not be homologous between these two groups.[64]
Further, ATP synthase, though homologous in all organisms, differs greatly between bacteria (including eukaryotic organelles such as mitochondria and chloroplasts) and the archaea/eukaryote nucleus group. The last common ancestor of all life (called LUCA) should have possessed an early version of this protein complex. As ATP synthase is obligate membrane bound, this supports the assumption that LUCA was a cellular organism. The RNA world hypothesis might clarify this scenario, as LUCA might have lacked DNA, but had an RNA genome built by ribosomes as suggested by Woese.[63]
A ribonucleoprotein world has been proposed based on the idea that oligopeptides may have been built together with primordial nucleic acids at the same time, which supports the concept of a ribocyte as LUCA. The feature of DNA as the material base of the genome might have then been adopted separately in bacteria and in archaea (and later eukaryote nuclei), presumably with the help of some viruses (possibly retroviruses as they could reverse transcribe RNA to DNA).[65]
See also
[edit]References
[edit]- ^ "procaryote". Merriam-Webster.com Dictionary. Merriam-Webster. Retrieved 2023-12-30.
- ^ a b "Prokaryotes: Single-celled Organisms". North Carolina State University.
- ^ Harper, Douglas. "prokaryote". Online Etymology Dictionary.
- ^ Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO (August 2005). "Protein structures forming the shell of primitive bacterial organelles". Science. 309 (5736): 936–8. Bibcode:2005Sci...309..936K. CiteSeerX 10.1.1.1026.896. doi:10.1126/science.1113397. PMID 16081736. S2CID 24561197.
- ^ Murat D, Byrne M, Komeili A (October 2010). "Cell biology of prokaryotic organelles". Cold Spring Harbor Perspectives in Biology. 2 (10): a000422. doi:10.1101/cshperspect.a000422. PMC 2944366. PMID 20739411.
- ^ Murat, Dorothee; Byrne, Meghan; Komeili, Arash (2010-10-01). "Cell Biology of Prokaryotic Organelles". Cold Spring Harbor Perspectives in Biology. 2 (10): a000422. doi:10.1101/cshperspect.a000422. PMC 2944366. PMID 20739411.
- ^ Wickstead B, Gull K (August 2011). "The evolution of the cytoskeleton". The Journal of Cell Biology. 194 (4): 513–25. doi:10.1083/jcb.201102065. PMC 3160578. PMID 21859859.
- ^ "Size scale of prokaryotic cells, from the largest to the smallest". Bionumbers. Retrieved 20 December 2024.
- ^ Schulz, H. N.; Jorgensen, B. B. (2001). "Big bacteria". Annual Review of Microbiology. 55: 105–137. doi:10.1146/annurev.micro.55.1.105. PMID 11544351.
- ^ Bauman RW, Tizard IR, Machunis-Masouka E (2006). Microbiology. San Francisco: Pearson Benjamin Cummings. ISBN 978-0-8053-7693-7.
- ^ Stoeckenius W (October 1981). "Walsby's square bacterium: fine structure of an orthogonal procaryote". Journal of Bacteriology. 148 (1): 352–60. doi:10.1128/JB.148.1.352-360.1981. PMC 216199. PMID 7287626.
- ^ Raven, Peter; Singer, Susan; Mason, Kenneth; Losos, Jonathan; Johnson, George (2013). "4. Cell structure: Prokaryotic cells". Biology. McGraw-Hill Education. p. 63. ISBN 978-0073383071.
- ^ Bobay, Louis-Marie (2020). The pangenome: Diversity, dynamics and evolution of genomes (PDF). Springer. pp. 253, 282. ISBN 978-3-030-38280-3.
- ^ a b c Chiang, Yin Ning; Penadés, José R.; Chen, John (2019). "Genetic transduction by phages and chromosomal islands: The new and noncanonical". PLoS pathogens. 15 (8): e1007878. doi:10.1371/journal.ppat.1007878. PMC 6687093. PMID 31393945.
- ^ a b Rosenshine I, Tchelet R, Mevarech M (September 1989). "The mechanism of DNA transfer in the mating system of an archaebacterium". Science. 245 (4924): 1387–9. Bibcode:1989Sci...245.1387R. doi:10.1126/science.2818746. PMID 2818746.
- ^ a b Patkowski, Jonasz (21 April 2023). "F-pilus, the ultimate bacterial sex machine". Nature Portfolio Microbiology Community.
- ^ a b Chen I, Dubnau D (March 2004). "DNA uptake during bacterial transformation". Nature Reviews Microbiology. 2 (3): 241–9. doi:10.1038/nrmicro844. PMID 15083159. S2CID 205499369.
- ^ Cabezón, Elena; Ripoll-Rozada, Jorge; Peña, Alejandro; de la Cruz, Fernando; Arechaga, Ignacio (2014). "Towards an integrated model of bacterial conjugation" (PDF). FEMS Microbiology Reviews. doi:10.1111/1574-6976.12085.
- ^ Solomon JM, Grossman AD (April 1996). "Who's competent and when: regulation of natural genetic competence in bacteria". Trends in Genetics. 12 (4): 150–155. doi:10.1016/0168-9525(96)10014-7. PMID 8901420.
- ^ Akamatsu T, Taguchi H (April 2001). "Incorporation of the whole chromosomal DNA in protoplast lysates into competent cells of Bacillus subtilis". Bioscience, Biotechnology, and Biochemistry. 65 (4): 823–9. doi:10.1271/bbb.65.823. PMID 11388459. S2CID 30118947.
- ^ Saito Y, Taguchi H, Akamatsu T (March 2006). "Fate of transforming bacterial genome following incorporation into competent cells of Bacillus subtilis: a continuous length of incorporated DNA". Journal of Bioscience and Bioengineering. 101 (3): 257–62. doi:10.1263/jbb.101.257. PMID 16716928.
- ^ Johnsborg O, Eldholm V, Håvarstein LS (December 2007). "Natural genetic transformation: prevalence, mechanisms and function". Research in Microbiology. 158 (10): 767–78. doi:10.1016/j.resmic.2007.09.004. PMID 17997281.
- ^ Fröls S, Ajon M, Wagner M, Teichmann D, Zolghadr B, Folea M, Boekema EJ, Driessen AJ, Schleper C, Albers SV (November 2008). "UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation" (PDF). Molecular Microbiology. 70 (4): 938–52. doi:10.1111/j.1365-2958.2008.06459.x. PMID 18990182. S2CID 12797510.
- ^ "Golden Dome Cave". National Park Service. 6November 2021. Archived from the original on 13 December 2022. Retrieved 11 February 2024.
{{cite web}}: Check date values in:|date=(help) - ^ Madigan T (2012). Brock biology of microorganisms (13th ed.). San Francisco: Benjamin Cummings. ISBN 9780321649638.
- ^ Costerton JW (2007). "Direct Observations". The Biofilm Primer. Springer Series on Biofilms. Vol. 1. Berlin, Heidelberg: Springer. pp. 3–4. doi:10.1007/978-3-540-68022-2_2. ISBN 978-3-540-68021-5.
- ^ Kaiser D (October 2003). "Coupling cell movement to multicellular development in myxobacteria". Nature Reviews. Microbiology. 1 (1): 45–54. doi:10.1038/nrmicro733. PMID 15040179. S2CID 9486133.
- ^ Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (October 1995). "Microbial biofilms". Annual Review of Microbiology. 49 (1): 711–745. doi:10.1146/annurev.mi.49.100195.003431. PMID 8561477.
- ^ Shapiro JA (1998). "Thinking about bacterial populations as multicellular organisms" (PDF). Annual Review of Microbiology. 52 (1): 81–104. doi:10.1146/annurev.micro.52.1.81. PMID 9891794. Archived from the original (PDF) on 2011-07-17.
- ^ Chua SL, Liu Y, Yam JK, Chen Y, Vejborg RM, Tan BG, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L (July 2014). "Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles". Nature Communications. 5 (1): 4462. Bibcode:2014NatCo...5.4462C. doi:10.1038/ncomms5462. PMID 25042103.
- ^ Balaban N, Ren D, Givskov M, Rasmussen TB (2008). "Introduction". Control of Biofilm Infections by Signal Manipulation. Springer Series on Biofilms. Vol. 2. Berlin, Heidelberg: Springer. pp. 1–11. doi:10.1007/7142_2007_006. ISBN 978-3-540-73852-7.
- ^ Costerton JW, Stewart PS, Greenberg EP (May 1999). "Bacterial biofilms: a common cause of persistent infections". Science. 284 (5418): 1318–22. Bibcode:1999Sci...284.1318C. doi:10.1126/science.284.5418.1318. PMID 10334980. S2CID 27364291.
- ^ Hogan CM (2010). "Extremophile". In Monosson E, Cleveland C (eds.). Encyclopedia of Earth. National Council of Science & the Environment.
- ^ Cobián Güemes, Ana Georgina; Youle, Merry; Cantú, Vito Adrian; Felts, Ben; Nulton, James; Rohwer, Forest (2016-09-29). "Viruses as Winners in the Game of Life". Annual Review of Virology. 3 (1). Annual Reviews: 197–214. doi:10.1146/annurev-virology-100114-054952. ISSN 2327-056X. PMID 27741409. S2CID 36517589.
- ^ Egel R (January 2012). "Primal eukaryogenesis: on the communal nature of precellular States, ancestral to modern life". Life. 2 (1): 170–212. Bibcode:2012Life....2..170E. doi:10.3390/life2010170. PMC 4187143. PMID 25382122.
- ^ Zimmer C (August 2009). "Origins. On the origin of eukaryotes". Science. 325 (5941): 666–8. doi:10.1126/science.325_666. PMID 19661396.
- ^ Brown JR (February 2003). "Ancient horizontal gene transfer". Nature Reviews. Genetics. 4 (2): 121–32. doi:10.1038/nrg1000. PMID 12560809. S2CID 22294114.
- ^ Forterre P, Philippe H (October 1999). "Where is the root of the universal tree of life?". BioEssays. 21 (10): 871–9. doi:10.1002/(SICI)1521-1878(199910)21:10<871::AID-BIES10>3.0.CO;2-Q. PMID 10497338.
- ^ Poole A, Jeffares D, Penny D (October 1999). "Early evolution: prokaryotes, the new kids on the block". BioEssays. 21 (10): 880–9. doi:10.1002/(SICI)1521-1878(199910)21:10<880::AID-BIES11>3.0.CO;2-P. PMID 10497339. S2CID 45607498.
- ^ Woese C (June 1998). "The universal ancestor". Proceedings of the National Academy of Sciences of the United States of America. 95 (12): 6854–9. Bibcode:1998PNAS...95.6854W. doi:10.1073/pnas.95.12.6854. PMC 22660. PMID 9618502.
- ^ Martin, William (2005). "Woe is the Tree of Life". In Sapp, Jan (ed.). Microbial Phylogeny and Evolution: Concepts and Controversies. Oxford: Oxford University Press. p. 139.
- ^ Carl Woese, J Peter Gogarten, "When did eukaryotic cells (cells with nuclei and other internal organelles) first evolve? What do we know about how they evolved from earlier life-forms?" Scientific American, October 21, 1999.
- ^ a b Hug, Laura A.; Baker, Brett J.; Anantharaman, Karthik; Brown, Christopher T.; Probst, Alexander J.; et al. (2016-04-11). "A new view of the tree of life". Nature Microbiology. 1 (5): 1–6. doi:10.1038/nmicrobiol.2016.48.
- ^ a b Stanier RY, Van Niel CB (1962). "The concept of a bacterium". Archiv für Mikrobiologie. 42 (1): 17–35. doi:10.1007/BF00425185. PMID 13916221. S2CID 29859498.
- ^ Chatton É (1937). Titres Et Travaux Scientifiques (1906-1937) De Edouard Chatton. Sète: Impr. E. Sottano.
- ^ Sapp, Jan (2005). "The Prokaryote-Eukaryote Dichotomy: Meanings and Mythology". Microbiology and Molecular Biology Reviews. 69 (2): 292–305. doi:10.1128/MMBR.69.2.292-305.2005. PMC 1197417. PMID 15944457.
- ^ Woese CR (March 1994). "There must be a prokaryote somewhere: microbiology's search for itself". Microbiological Reviews. 58 (1): 1–9. doi:10.1128/MMBR.58.1.1-9.1994. PMC 372949. PMID 8177167.
- ^ Sapp J (June 2005). "The prokaryote-eukaryote dichotomy: meanings and mythology". Microbiology and Molecular Biology Reviews. 69 (2): 292–305. doi:10.1128/MMBR.69.2.292-305.2005. PMC 1197417. PMID 15944457.
- ^ a b Coté G, De Tullio M (2010). "Beyond Prokaryotes and Eukaryotes: Planctomycetes and Cell Organization". Nature.
- ^ Bruce Alberts; et al. (2002). The Molecular Biology of the Cell (fourth ed.). Garland Science. p. 808. ISBN 0-8153-3218-1.
- ^ Thanbichler M, Wang SC, Shapiro L (October 2005). "The bacterial nucleoid: a highly organized and dynamic structure". Journal of Cellular Biochemistry. 96 (3): 506–21. doi:10.1002/jcb.20519. PMID 15988757. S2CID 25355087.
- ^ Helinski DR (December 2022). Kaper JB (ed.). "A Brief History of Plasmids". EcoSal Plus. 10 (1): eESP00282021. doi:10.1128/ecosalplus.ESP-0028-2021. PMC 10729939. PMID 35373578.
- ^ Johnston C, Caymaris S, Zomer A, Bootsma HJ, Prudhomme M, Granadel C, Hermans PW, Polard P, Martin B, Claverys JP (2013). "Natural genetic transformation generates a population of merodiploids in Streptococcus pneumoniae". PLOS Genetics. 9 (9): e1003819. doi:10.1371/journal.pgen.1003819. PMC 3784515. PMID 24086154.
- ^ a b Harold FM (June 1972). "Conservation and transformation of energy by bacterial membranes". Bacteriological Reviews. 36 (2): 172–230. doi:10.1128/MMBR.36.2.172-230.1972. PMC 408323. PMID 4261111.
- ^ Shih YL, Rothfield L (September 2006). "The bacterial cytoskeleton". Microbiology and Molecular Biology Reviews. 70 (3): 729–754. doi:10.1128/MMBR.00017-06. PMC 1594594. PMID 16959967.
- ^ Michie KA, Löwe J (2006). "Dynamic filaments of the bacterial cytoskeleton" (PDF). Annual Review of Biochemistry. 75 (1): 467–92. doi:10.1146/annurev.biochem.75.103004.142452. PMID 16756499. Archived from the original (PDF) on November 17, 2006.
- ^ Fuerst JA (2005). "Intracellular compartmentation in planctomycetes". Annual Review of Microbiology. 59 (1): 299–328. doi:10.1146/annurev.micro.59.030804.121258. PMID 15910279.
- ^ Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP (2013). "Three-dimensional reconstruction of bacteria with a complex endomembrane system". PLOS Biology. 11 (5): e1001565. doi:10.1371/journal.pbio.1001565. PMC 3660258. PMID 23700385.
- ^ Kempes, Christopher P.; Dutkiewicz, Stephanie; Follows, Michael J. (10 January 2012). "Growth, metabolic partitioning, and the size of microorganisms" (PDF). Proceedings of the National Academy of Sciences. 109 (2): 495–500. doi:10.1073/pnas.1115585109. PMC 3258615. PMID 22203990.
- ^ a b c Castelle CJ, Banfield JF (March 2018). "Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life". Cell. 172 (6): 1181–1197. doi:10.1016/j.cell.2018.02.016. PMID 29522741.
- ^ a b Mattiroli F, Bhattacharyya S, Dyer PN, White AE, Sandman K, Burkhart BW, Byrne KR, Lee T, Ahn NG, Santangelo TJ, Reeve JN, Luger K (August 2017). "Structure of histone-based chromatin in Archaea". Science. 357 (6351): 609–612. Bibcode:2017Sci...357..609M. doi:10.1126/science.aaj1849. PMC 5747315. PMID 28798133.
- ^ Cavalier-Smith T (March 2002). "The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa". International Journal of Systematic and Evolutionary Microbiology. 52 (Pt 2): 297–354. doi:10.1099/00207713-52-2-297. PMID 11931142. Archived from the original on 2017-07-29. Retrieved 2019-03-21.
- ^ a b Lane N (2015). "Energy, Evolution, and the Origins of Complex Life". The Vital Question. W. W. Norton. p. 77. ISBN 978-0-393-08881-6.
- ^ a b Barry ER, Bell SD (December 2006). "DNA replication in the archaea". Microbiology and Molecular Biology Reviews. 70 (4): 876–887. doi:10.1128/MMBR.00029-06. PMC 1698513. PMID 17158702.
- ^ Forterre P (2006). "Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain". PNAS. 103 (10): 3669–3674. Bibcode:2006PNAS..103.3669F. doi:10.1073/pnas.0510333103. PMC 1450140. PMID 16505372.
External links
[edit]- Prokaryote versus eukaryote, BioMineWiki Archived 2012-10-25 at the Wayback Machine
- The Taxonomic Outline of Bacteria and Archaea
- The Prokaryote-Eukaryote Dichotomy: Meanings and Mythology
- Quiz on prokaryote anatomy
- TOLWEB page on Eukaryote-Prokaryote phylogeny
![]() This article incorporates public domain material from Science Primer. NCBI. Archived from the original on 2009-12-08.
This article incorporates public domain material from Science Primer. NCBI. Archived from the original on 2009-12-08.